Application, interpretability, and clinical translation of deep learning algorithms for medical images
My research reduces dependence on specialized medical imaging devices, biological and chemical processes and creates new paradigms for using low-cost images captured using simple optical principles for point-of-care clinical diagnosis. My research lab has demonstrated a series of generative, prediction and classification algorithms for obtaining medical diagnostic information of organs and tissues from simple images captured by low-cost devices. For example:
- Generalizability of deep learning models for segmenting complex patterns from images are not well understood and are based on anecdotal assumptions that increasing training data improved performance. In a Cell Reports Methods paper my lab reported an end-to-end toolkit for improving generalizability and transparency of clinical-grade DL architectures [1]. Researchers and clinicians can identify hidden patterns embedded in images and overcome under specification of key non-disease and clinical labels causal to decreasing false-positive or negative outcomes in high-dimensional learning systems. The key findings from this study focus on the evaluation of medical images, but these methods and approach should generalize to all other RGB and gray scale natural-world image segmentations. Methods for benchmarking, visualization, and validation of deep learning models and images communicated in this study have wide applications in biomedical research and uncertainty estimations for regulatory science purposes [1, 2].
- In collaboration with Brigham and Women’s Hospital in Boston, MA, my research lab devised and published a novel “Computational staining” system to digitally stain photographs of unstained tissue biopsies with Haematoxylin and Eosin (H\&E) dyes to diagnose cancer [3]. This research also described an automated “Computational destaining” algorithm that can remove dyes and stains from photographs of previously stained tissues, allowing reuse of patient samples. The methods communicated in this study use neural networks to help physicians provide timely information about the anatomy and structure of the organ and saving time and precious biopsy samples.
- In collaboration with Stanford University School of Medicine and Harvard Medical School I led research studies that reported several novel mechanistic insights and methods to facilitate benchmarking and clinical and regulatory evaluations of generative neural networks and computationally H\&E-stained images. Specifically, high fidelity, explainable, and automated computational staining and destaining algorithms to learn mappings between pixels of nonstained cellular organelles and their stained counterparts were trained. A novel and robust loss function was devised for the deep learning algorithms to preserve tissue structure. The virtual staining neural network models developed by my lab were generalizable to accurately stain previously unseen images acquired from patients and tumor grades not part of training data. Neural activation maps in response to various tumors and tissue types were generated to provide the first instance of explainability and mechanisms used by deep learning models for virtual H\&E staining and destaining. And image processing analytics and statistical testing were used to benchmark the quality of generated images. Finally, I evaluated computationally stained images for prostate tumor diagnoses with multiple pathologists for clinical evaluations [4].
- More recently, my research lab communicated a deep weekly-supervised learning GBP algorithm and Resnet-18 classifier to localize class-specific (tumor) regions on virtually stained images generated from GAN-CS model [5].
- In collaboration with Beth Israel Deaconess Medical Center in Boston, MA, my research lab investigated the use of dark field imaging of capillary bed under the tongue of consenting patients in emergency rooms for diagnosing sepsis (a blood borne bacterial infection). A neural network capable of distinguishing between images from non-septic and septic patients with more than 90% accuracy was reported for the first time [6]. This approach can rapidly stratify patients, offer rational use of antibiotics, and reduce disease burden in hospital emergency rooms and combat antimicrobial resistance.
- I led research studies with neural networks that successfully predicted signatures associated with fluorescent porphyrin biomarkers (linked with tumors and periodontal diseases) from standard white-light photographs of the mouth, thus reducing the need for fluorescent imaging [7].
- My lab has communicated research studies reporting automated segmentation of oral diseases from standard photographs [8] by neural networks and correlations with systemic health conditions such as optic nerve abnormalities in patients for personalized risk scores [9].
Examples listed above describe my labs contributions to design novel neural networks and processes that can assist physicians and patients by next-generation computational medicine algorithms at the point-of-care which integrate seamlessly into clinical workflows in hospitals all over the world.
Peer-Reviewed Publications
-
A deep-learning toolkit for visualization and interpretation of segmented medical images. 2021. [Abstract] [Full Paper]
Sambuddha Ghosal and Shah P*
( *Senior author supervising research)
Cell Reports Methods 1, 100107, 2021
-
Uncertainty quantified deep learning for predicting dice coefficient of digital histopathology image segmentation. 2021. [arXiv Preprint]
Sambuddha Ghosal, Xie A and Shah P*
( *Senior author supervising research)
arXiv:2011.05791 [stat.ML]
-
Computational histological staining and destaining of prostate core biopsy RGB images with generative adversarial neural networks. 2018. [Abstract] [Full Paper]
Aman Rana, Yauney G, Lowe A, Shah P*
(*Senior author supervising research)
17th IEEE International Conference of Machine Learning and Applications. DOI: 10.1109/ICMLA.2018.00133
-
Use of deep learning to develop and analyze computational hematoxylin and eosin staining of prostate core biopsy images for tumor diagnosis. 2020. [Abstract] [Full Paper]
Aman Rana, Lowe A, Lithgow M, Horback K, Janovitz T, Da Silva A, Tsai H, Shanmugam V, Bayat A, Shah P*
(*Senior author supervising research)
JAMA Network. DOI: 10.1001/jamanetworkopen.2020.5111
-
Automated end-to-end deep learning framework for classification and tumor localization from native non-stained pathology images. 2021. [Abstract] [Full Paper]
Akram Bayat, Anderson C, Shah P*♯
(*Senior author supervising research, ♯Selected for Deep-dive spotlight session)
SPIE Proceedings. DOI: 10.1117/12.2582303
-
Machine learning algorithms for classification of microcirculation images from septic and non-septic patients. 2018. [Abstract] [Full Paper]
Perikumar Javia, Rana A, Shapiro NI, Shah P*
(*Senior author supervising research)
17th IEEE International Conference of Machine Learning and Applications. DOI: 10.1109/ICMLA.2018.00097
-
Convolutional neural network for combined classification of fluorescent biomarkers and expert annotations using white light images. 2017. [Abstract] [Full Paper]
Gregory Yauney, Angelino K, Edlund D, Shah P*♯
(*Senior author supervising research, ♯Selected for oral presentation)
17th annual IEEE International Conference on BioInformatics and BioEngineering. DOI: 10.1109/BIBE.2017.00-37
-
Automated segmentation of gingival diseases from oral images. 2017. [Abstract] [Full Paper]
Aman Rana, Yauney G, Wong L, Muftu A, Shah P*
(*Senior author supervising research)
IEEE-NIH 2017 Special Topics Conference on Healthcare Innovations and Point-of-Care Technologies. DOI: 10.1109/HIC.2017.8227605
-
Automated process incorporating machine learning segmentation and correlation of oral diseases with systemic health. 2019. [Abstract] [Full Paper]
Gregory Yauney, Rana A, Javia P, Wong L, Muftu A, Shah P*
(*Senior author supervising research)
41st IEEE International Engineering in Medicine and Biology Conference. DOI: 10.1109/EMBC.2019.8857965
Select Talks
- 2021 - Interpretable AI and Explainable AI
- 2020 - Novel deep learning systems for oncology imaging, digital medicines and real world data
- 2019 - Understanding biomarker science: from molecules to images
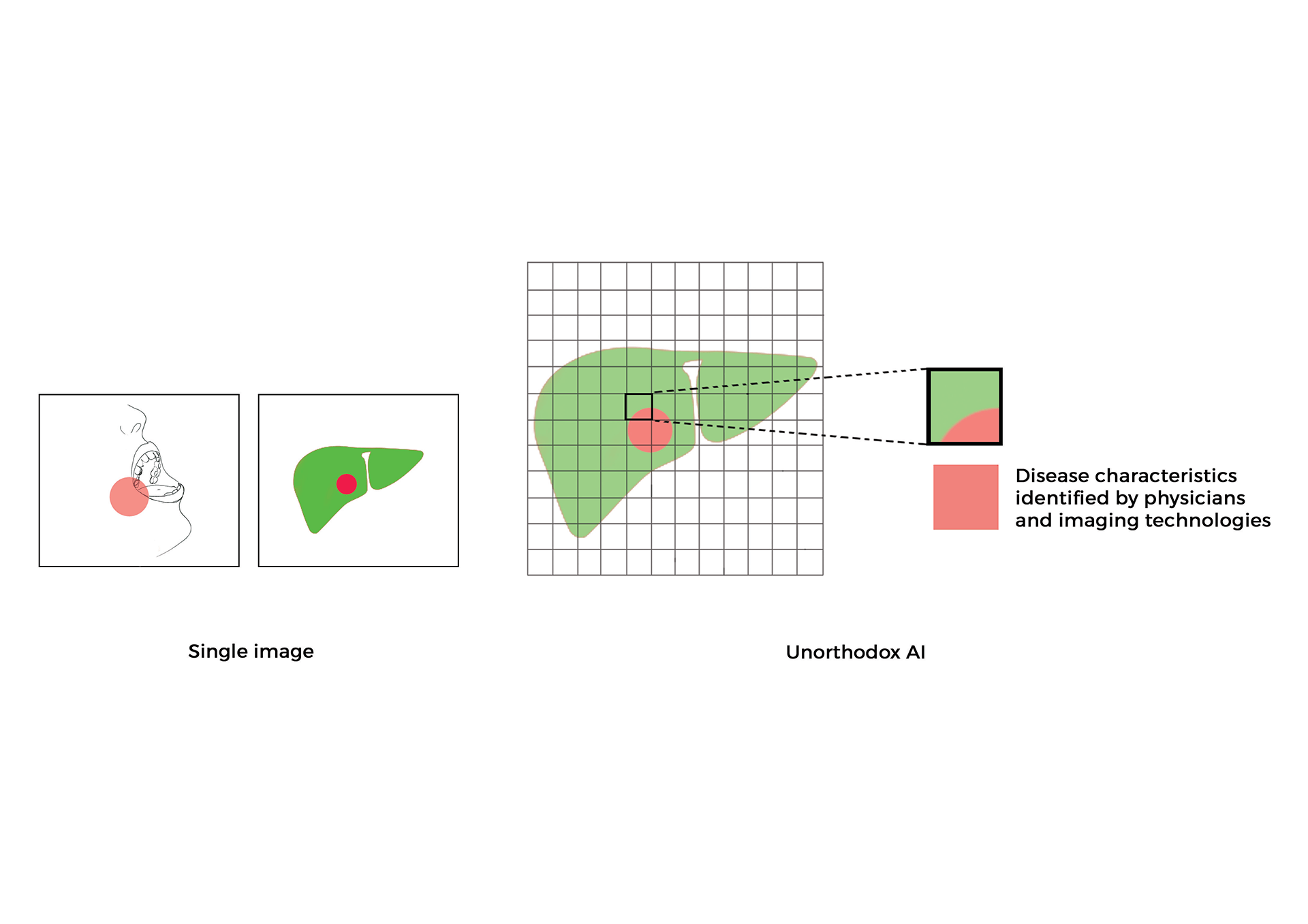
- 2019 - Unorthodox AI and machine learning use cases and future of clinical medicine
- 2019 - Machine learning methods for biomedical image analysis
- 2018 - Novel machine learning and pragmatic computational medicine approaches to improve health outcomes for patients
- 2018 - TED Talk - How AI is making it easier to diagnose disease
- 2017 - Artificial intelligence for medical images
Press
- 2020 - Deep learning accurately stains digital biopsy slides
- 2017 - New visions for the world we know: Notes from an early morning of TED Fellows talks
- 2017 - Innovation and Inspiration From TEDGlobal 2017
- 2017 - TED: Phones and drones transforming healthcare
Honors
- 2017 - TED Fellow